I was lucky to get one of the ERC grants in 2020, meaning that my work will be supported by 1,7 million euro. The work will focus on dryland trees in satellite images. Here is the abstract: Drylands cover approximately 65 million km² of the Earth’s land surface but their tree and shrub cover is … Continue reading 1.7 million € from an ERC Starting Grant
Author: Martin Brandt
Closed – PhD Fellow in Assessing Trees in African Drylands
The Department of Geosciences and Natural Resource Management in Faculty of Science at the University of Copenhagen is inviting applications for a PhD position in assessing tree cover in African drylands using very high resolution satellite imagery and deep learning. The position will start on October 1st 2020 or as soon as possible hereafter and … Continue reading Closed – PhD Fellow in Assessing Trees in African Drylands
Closed – Open PhD position
University of Copenhagen, Kayrros, Laboratoire des Sciences du Climat et de l’Environnement (LSCE), INRAE Bordeaux are looking for a PhD candidate who will be employed by LSCE and mainly based at the Department of Geosciences and Natural Resource Management (IGN) in Copenhagen, Denmark. PhD topic Dynamic forest carbon maps from very high resolution satellite data … Continue reading Closed – Open PhD position
New study in Nature Communications
Land use policies have turned southern China into one of the most intensively managed forest regions in the world, with actions maximizing forest cover on soils with marginal agricultural potential while concurrently increasing livelihoods and mitigating climate change. Based on satellite observations, here we show that diverse land use changes in southern China have increased … Continue reading New study in Nature Communications
New paper in Nature Plants
We have a new paper online showing aboveground biomass carbon fluxes in the global tropics using the SMOS L-VOD data. https://www.nature.com/articles/s41477-019-0478-9
New papers on the impact of changed rainfall patterns on savanna vegetation
Climate change includes not only changed rainfall amounts, but also the distribution, intensity and frequency of heavy rainfall events changes. This leads to a a generally decreased relationship between annual rainfall and vegetation production. Also, herbaceous and woody vegetation benefit differently from changed rainfall patterns. We have now 2 new studies online, exploring how changed … Continue reading New papers on the impact of changed rainfall patterns on savanna vegetation
Tree cover promoted in semi-arid Sahelian farms – new publication in Nature Geoscience
More people equal more trees in semi-arid West Africa - Our new study published in Nature Geoscience questions ‘received wisdom’ as concerns the relationship between human agency and woody vegetation of West Africa. We demonstrate that in low-rainfall areas woody cover is denser in cultivated areas than in savannas, and close to settlements rather that … Continue reading Tree cover promoted in semi-arid Sahelian farms – new publication in Nature Geoscience
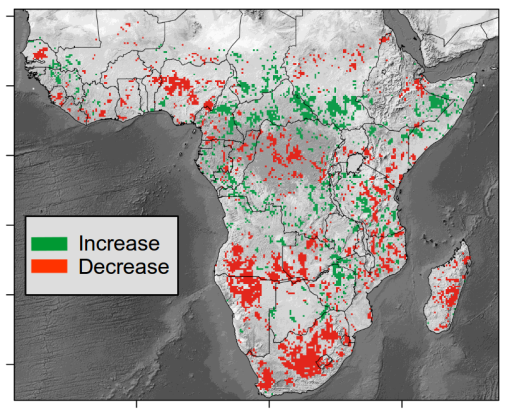
A new tool to monitor aboveground vegetation carbon stocks: first application to the African continent
Our new study uses unprecedented data sources to measure vegetation carbon stock dynamics at continental scale. The study demonstrates that over the African continent, the net carbon balance is negative for 2010-2016, and that most of the carbon losses occurred in dryland savannahs. The results were published in the journal Nature Ecology and Evolution. The … Continue reading A new tool to monitor aboveground vegetation carbon stocks: first application to the African continent
Why our research matters
At some point, many scientists working in geographic fields may have asked themselves if there is any "use" in the work we do. The main aim of a study should not be to get it published in a high impact journal but to share research results with as many people as possible. However, here comes … Continue reading Why our research matters
New Publication in Nature Sustainability
Now this is something really cool: -We have a paper in the first ever issue of the new journal Nature Sustainability -Both the cover of the first issue and the website banner are my photos I shot last summer in Southern China -The paper is the thesis of my girlfriend and it is the cover … Continue reading New Publication in Nature Sustainability