Here's a simple way of processing MODIS: downloading, extracting the bands from HDF as GEOTIFF, merging tiles and reprojecting. Area extraction, NDVI calculation, cloud filtering and more can be done in GRASS for example. It's Linux based, but can easily used in Windows. My way might not be the most elegant, but it's fast and … Continue reading Downloading and processing MODIS with fast and free tools
Tag: time series
Recent woody vegetation trends in Sahel
Our new paper looks at recent dynamics in woody vegetation in Sahel and finds some interesting patterns which are mainly controlled by human population density. Martin Brandt, Pierre Hiernaux, Kjeld Rasmussen, Cheikh Mbow, Laurent Kergoat, Torbern Tagesson, Yahaya Ibrahim, Abdoulaye Wele, Compton J. Tucker, Rasmus Fensholt. Assessing woody vegetation trends in Sahelian drylands using MODIS … Continue reading Recent woody vegetation trends in Sahel
Smoothing/Filtering a NDVI time series using a Savitzky Golay filter and R
Coarse scaled NDVI (or FAPAR, EVI...) images have a high temporal frequency and are delivered as Maximum Value Composites (MVC) of several days, which means the highest value is taken, assuming that clouds and other contaminated pixels have low values. However, especially in areas with a rainy season, the composites over 10-16 days still contain … Continue reading Smoothing/Filtering a NDVI time series using a Savitzky Golay filter and R
Pixel-wise regression between two raster time series (e.g. NDVI and rainfall)
Doing a pixel-wise regression between two raster time series can be useful for several reasons, for example: find the relation between vegetation and rainfall for each pixel, e.g. a low correlation could be a sign of degradation derive regression coefficients to model the depending variable using the independend variable (e.g. model NDVI with rainfall data) … Continue reading Pixel-wise regression between two raster time series (e.g. NDVI and rainfall)
Environmental change in time series – An interdisciplinary study in the Sahel of Mali and Senegal
Our next article is now online in the Journal of Arid Environments: Brandt, M., Romankiewicz, C., Spiekermann, R. & C. Samimi: Environmental change in time series – An interdisciplinary study in the Sahel of Mali and Senegal. Journal of Arid Environments 105, June 2014, Pages 52-63. It is available here: http://www.sciencedirect.com/science/article/pii/S0140196314000536 Abstract: Climatic changes and human activities have caused … Continue reading Environmental change in time series – An interdisciplinary study in the Sahel of Mali and Senegal
Renaming, converting, clipping: script based raster time series processing
When working with remotely sensed time series data (e.g. MODIS, AVHRR, GIMMS, etc.), bulk processing can save a lot of time. Using a terminal in a Linux environment, simple scripts can process thousends of files in short time. Here are some basic hints on how to start, gdal has to be installed. For Windows users … Continue reading Renaming, converting, clipping: script based raster time series processing
Pixel-wise time series trend anaylsis with NDVI (GIMMS) and R
The GIMMS dataset is currently offline and the new GIMMS3g will soon be released, but it does not really matter which type of data is used for this purpose. It can also be SPOT VGT, MODIS or anything else as long as the temporal resolution is high and the time frame is long enough to … Continue reading Pixel-wise time series trend anaylsis with NDVI (GIMMS) and R
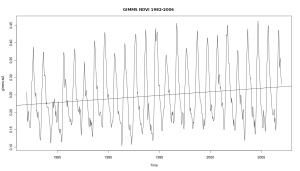
Simple time series analysis with GIMMS NDVI and R
Time series analysis with satellite derived greenness indexes (e.g. NDVI) is a powerfull tool to assess environmental processes. AVHRR, MODIS and SPOT VGT provide global and daily imagery. Creating some plots is a simple task, and here is a rough start how it is done with GIMMS NDVI. All we need is the free software … Continue reading Simple time series analysis with GIMMS NDVI and R
Converting GPCC gridded rainfall 1901-2010@0.5° to monthly Geotiffs
GPCC rainfall data contains the largest database worldwide, with over 90 000 weather-stations interpolated to a 0.5° grid. It includes S. Nicholsons dataset for Africa and is thus a good source for gridded monthly precipitation. It covers the period 1901-2010, however, it comes in a strange dataformat and it is a long way until we get … Continue reading Converting GPCC gridded rainfall 1901-2010@0.5° to monthly Geotiffs
Detecting environmental change using time series, high resolution imagery and field work – a case study in the Sahel of Mali
Climatic changes and population pressure have caused major environmental change in the Sahel during the last fifty years. Many studies use coarse resolution NDVI time series such as GIMMS to detect environmental trends; however explanations for these trends remain largely unknown.We suggest a five-step methodology for the validation of trends with a case study on the Dogon Plateau, … Continue reading Detecting environmental change using time series, high resolution imagery and field work – a case study in the Sahel of Mali